
The most highly used MD program in recent years is the GROMACS molecular simulation toolkit. GROMACS is cited by ~2,500 scientific articles per year, a rate that has been rapidly increasing recently. GROMACS is known for its high performance and its efficient, effective use of system resources, but new, massively parallel HPC (high-performance computing) architectures present a continuing challenge. At CMB, we are helping to meet this challenge. Specifically, we are working to upgrade GROMACS’ threading framework to keep all CPU cores busy by overlapping computing and communication. This requires careful rescheduling of threads to different tasks during data transfers, which was previously time that threads spent idle. This must be done while avoiding thread-management overhead, which can erode performance gains. Finally, all this has to be done within the narrow two millisecond window when GROMACS computes atom positions, forces, etc. for the next simulation step. To meet this goal, we are developing a new threading framework, called STS (static thread scheduler) (https://github.com/eblen/sts) and integrating it into GROMACS. Funding provided through the INTEL(R) IPCC Program.

The performance of multimillion-atom MD simulations on petascale supercomputers is limited by the most commonly used electrostatic method: PME. Particle mesh ewald (PME) requires two 3D fast fourier transforms (FFT) and these in turn require global communication. These global communications are very time consuming on massively parallel supercomputers, such as Jaguar at ORNL. Our recent paper shows that for the chosen test system (lignocellulosic biomass) the reaction field (RF) method produces very accurate results when compared to results obtained using PME. As seen in the graph, a simulation using RF shows significant better scaling than one of the same system using PME.
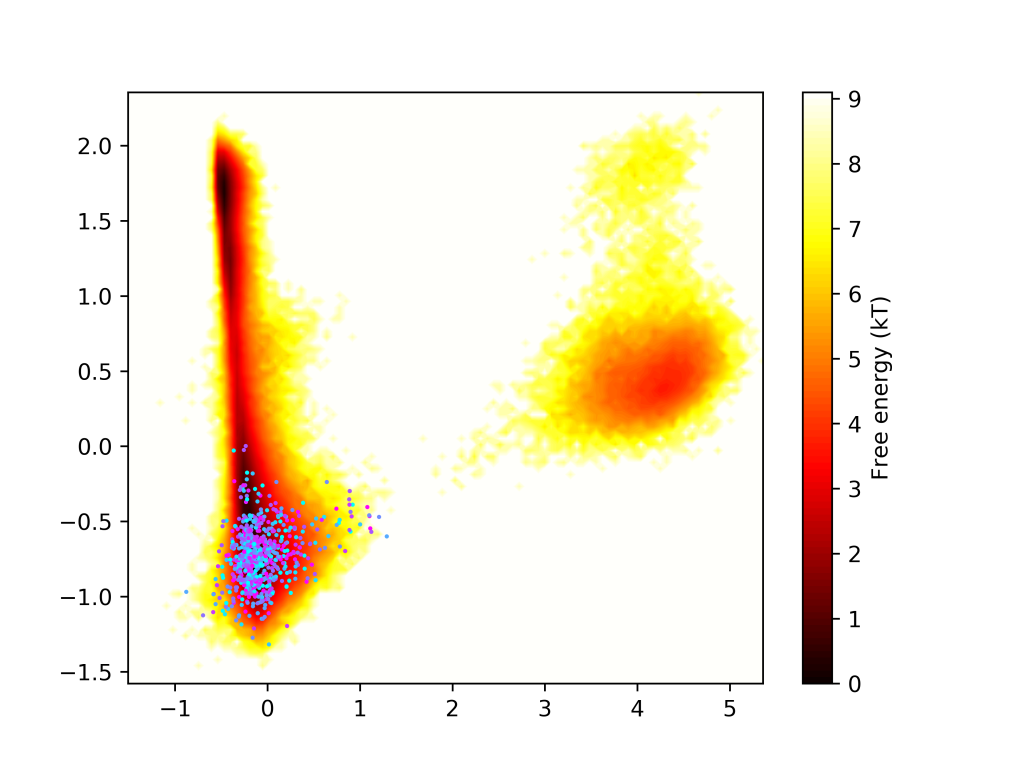
We are working on a software package that automates biomolecular dynamics simulation workflows. With AdaptiveMD, you can create workflows that iteratively restart a swarm of simulation replicates by sampling initial states from a Markov State Model of pre-existing data. Look at the github page for instructions on how to install and use the package.