
Christy Franklin Wins 2025 Administrative Excellence Award
Christy Franklin has been awarded the 2025 Biosciences Division Distinguished Achievement Award for Administrative Excellence! Christy has been our administrative

Christy Franklin has been awarded the 2025 Biosciences Division Distinguished Achievement Award for Administrative Excellence! Christy has been our administrative

One of Jeremy’s ex graduate-students, Frauke Graeter, is taking up a Directorship at the Max Planck Institute for Polymer Research
We are proud to announce that Center for Molecular Biophysics, along with the Chemical Sciences Division, University of Tennessee, and

We are pleased to announce that former CMB Postdoc Vaidyanathan M.S. has accepted an offer from Oak Ridge National Laboratory

We are thrilled to announce that Madhulika Gupta, a former CMB Postdoc, has been nominated by the Anusandhan National Research

East Tennessee State University has recently granted Joshua Olaf Aggrey, a graduate student researcher at ETSU, the 2024 Appalachian Student

We are thrilled to have Mohan on our team at CMB, where he has already made significant contributions during his

Congratulations to Adam Green, who won first prize for best poster at the 2019 Gordon Conference on Multidrug Efflux Systems.
Frank Noe, who is a close collaborator and was a PhD student with Jeremy Smith in Heidelberg, has received the 2019 Early Career Award in Theoretical Chemistry of the American Chemical Society (ACS).

Congratulations to former postdoc Petra Imhof for accepting a tenured professorship in chemistry at the University of Stavanger, Norway!

Michael Galloway has received an award from the Department of Energy (DOE) and the Department of Veterans Affairs (VA) in recognition of his exceptional service towards DOE-VA partnership. Mr. Galloway has provided great efforts in building and supporting the VA enclave at Oak Ridge National Laboratory, enabling VA and DOE researchers from across US to work in collaborative way.
Congratulations Michael!
Click here for details:
Congratulations to Mafalda Nina, one of Jeremy Smith’s first Ph.D. students, who has been named Global R&D Portfolio Manager and Head of Innovation Programs at Syngenta, one of the world’s biggest biotechnology companies.
For details, check out the link: https://www.youtube.com/watch?v=VLDO2DyFnIc
Best poster award (3rd) at 6th Annual Research Symposium: Oak Ridge Postdoctoral Association (ORPA), Oak Ridge National Laboratory, TN
Center for Molecular Biophysics scientist Jerry Parks along with several other ORNL scientist recently had their work published in Nature Communications. The following presses releases discuss the article:
“JC Gumbart and his team have used Titan, the Cray XK7 supercomputer at the Oak Ridge Leadership Computing Facility, a Department of Energy (DOE) user facility, to simulate the shape and related stability of AcrA as it interacts with other efflux pump components. Gumbart and his team gathered a trove of data in just one year. “It would have taken at least four to five years to obtain using common supercomputing resources,” he says. Team members include Jerry Parks of Oak Ridge National Laboratory; Jerome Baudry of the University of Alabama, Huntsville; Helen Zgurskaya of the University of Oklahoma; University of Tennessee, Knoxville graduate student Adam Green; and Georgia Tech graduate student Anthony Hazel.”
Former member Sally Ellingson used Big Data and Machine Learning to Improve Drug Development.
The research teams of L. Darryl Quarles, MD, of the University of Tennessee Health Science Center (UTHSC), Jeremy C. Smith, PhD, of the University of Tennessee, Knoxville (UTK) and Oak Ridge National Laboratory (ORNL), and Jerome Baudry, PhD (who was at UTK, but has since moved to the University of Alabama in Huntsville) have discovered which receptor key chemicals found in green tea interact with, according to work published in the article, “GPCR6A is a Molecular Target for the Natural Products Gallate and EGCG in Green Tea,” in the journal Molecular Nutrition and Food Research, a peer-reviewed scientific journal that covers the latest research in nutrition and food science.”-taken from the link.
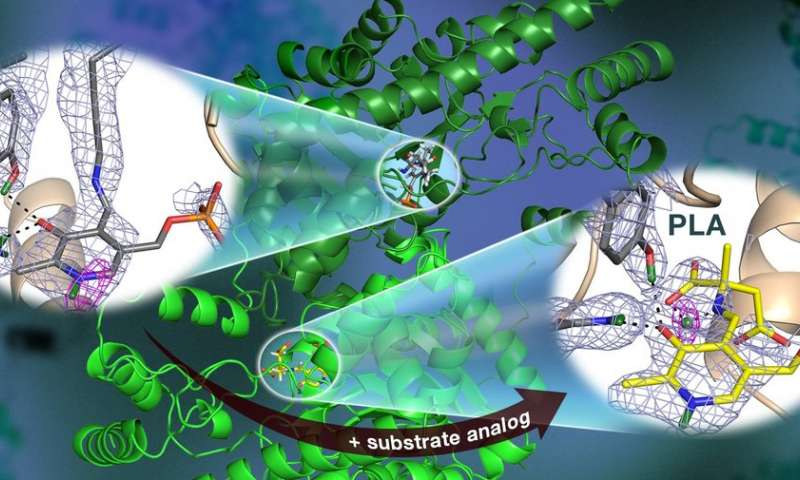
Researchers from Department of Energy’s ORNL used neutron crystallography to investigate human carbonic anhydrase II (hCA II) and its interaction with three glaucoma drugs. The study was published in the journal Structure.

The article titled: “The Title-dependent potential of mean force of a pair of DNA oligomers from all-atom moecular dynamics simulations” was selected as under the category of Soft matter, biophysics and liquids.
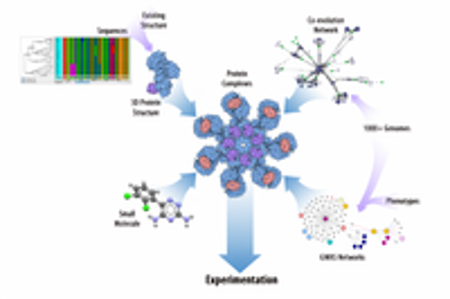
A multi-disciplinary team from Oak Ridge National Laboratory will be using TITAN and other super computer to study Populus trichocarpa (black cottonwood), which has potential in biofuels production.
Ada Sedova, a postdoctoral researcher at the OLCF who works with researchers here at CMB has been helping to bridge the gap between experiments in the lab and computation using supercomputers.
Former member Ruggero Cortini recently published an article as an Emerging leader and was interviewed by the Journal of Physics: Condensed Matter.
The BioEnergy Science Center led by Oak Ridge National Laboratory and a team of researchers including Jeremy Smith has ended after 10 years. One goal of the center was to improve biofuels production and the license to Enchi Corporation reached this goal.

According Almetric.com the paper titled “Mechanism of lignin inhibition of enzymatic biomass deconstruction” published in Biotechnology for Biofuels in listed as one the top influential papers of 2016. The article was written by Josh Vermaass, former member of the Center for Molecular Biophysics.
Director of the Center of Molecular Biophysics , Jeremy Smith, along with Howard Hall, another UT governor’s chair discuss the need for research to counteract chemical attacks.
Our very own Sarah Cooper has received the National Science Foundation Graduate Research fellowship. This award is for a three year period.
Researchers at the University of Oklahoma and Oak Ridge National Laboratory worked together to discover a new class of drugs that could combat antibiotic resistance. CMB group members at UT and ORNL worked with scientists at the University of Oklahoma and St. Louis University to discover molecules that combat antibiotic resistance. The team combined supercomputer simulations on Titan with laboratory experiments to discover molecules that disrupt the structure and function of the AcrAB-TolC multidrug efflux pump in E. coli, restoring the ability of existing antibiotics to kill the bacteria.
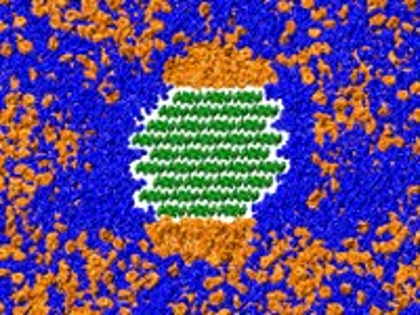
Researchers at ORNL uncover the chemical reaction that helps break down biomass for biofuel. Barmak Mostofian has published his work on biomass pretreatment in the Journal of the American Chemical Society. In this recent interview, Micholas Smith and Xiaolin Cheng comment on the novelty and the impact of this study.
The Center for Molecular Biophysics uses the next-generation Intel Xeon Phi processors to perform molecular dynamics research.
A group of three teachers, came to CMB to study molecular dynamics. Hector Vazquez, John Eben, and Adam Green assisted the teachers and gave talks during the visit.
Researchers at the University of Oklahoma and Oak Ridge National Laboratory worked together to discover a new class of drugs that could combat antibiotic resistance. CMB group members at UT and ORNL worked with scientists at the University of Oklahoma and St. Louis University to discover molecules that combat antibiotic resistance. The team combined supercomputer simulations on Titan with laboratory experiments to discover molecules that disrupt the structure and function of the AcrAB-TolC multidrug efflux pump in E. coli, restoring the ability of existing antibiotics to kill the bacteria.
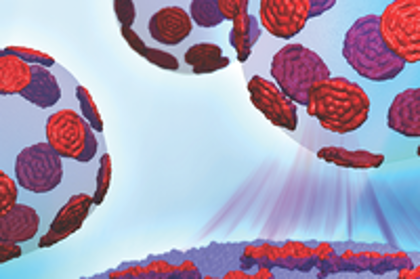
CMB researcher Xiolin Cheng, along with SNS researcher John Katsaras use Titan and the beam lines at SNS to study lipid rafts.
In a recent article in Scientific Computing the use of GROMACS by Jeremy Smith and team is highlighted. The article discusses all the research areas GROMACS is used at the Center for Molecular Biophysics.

Xiaohu’s recent article in Nature Physics about protein dynamics and movement is highlighted in the To the Point section.
Jerry Park’s work on mercury is discussed in the article “Focus on Mercury”
The Focus on Health article this month is about Jerome Baudry’s research using supercomputers to test new clinical drugs at a cellular level.
“Lipi Thukral, a computational structural biologist at the IGIB, sees the DBT’s goal of creating a genomics hub as an opportunity for India to enter the emerging field of precision medicine, which uses genomic, physiological and other data to tailor treatments to the individual. Achieving that will require clinicians to gather extensive data and to interact better with academics to analyse the data, says Thukral. To succeed, the strategy will also need a feedback mechanism between the biotech corporate sector and academic institutions, policies to protect scientists undertaking high-risk, high-reward research and a reorientation of academia towards more industry-relevant research, she adds.”
Jeremy Smith’s team test the pretreatment Cosolvent Enhanced Lignocellulose Fraction (CELF) using supercomputer TITAT and molecular dynamics coded GROMACS. The team discovered CELF works because water and tetrahydrofuran (THF) binds to lignin and creates a barrier allowing for easier removal of lignin. This discovery will hopefully lead to further research.
Jeremy’s Smith team uses supercomputer TITAN and the molecular dynamics code GROMACS to run simulations explaining why lignin is such a problem for cost-effective ethanol. Along the way they refined a process for analyzing large-scale simulations using parallel analysis.
Liang Hong’s work on understanding the different types of protein motion by combining dynamic neutron scattering experiments with molecular dynamics computer simulations has been highlighted on the Department of Energy’s Office of Science page.
The Department of Energy has awarded 100 million processor hours on its leadership computing facilities for our research project “A Generic Plant Cell Wall and its Deconstruction for Bioenergy.”
Congratulations to Loukas Petridis, Xiaolin Cheng, and Jeremy Smith!
Jerry Parks comments on the combination of high-performance computing with neutron-diffraction experiments to understand the acid/base chemistry of enzymatic biomass hydrolysis in this recent interview.
Jeremy Smith comments on the synergy in scientific research between the University of Tennessee and the Oak Ridge National Lab on NPR.
Jeremy has been interviewed for the 40th anniversary of the European Molecular Biology Laboratory (EMBL). He is an EMBL alumni, who did his PhD work at the Grenoble location in France.
Read here:
EMBL
Our Biofuels project is highlighted in the news.
Christopher Carmona, an undergraduate student in our lab, studied the effect of genetically modified lignin on plant cell wall structure.
Read here:
OLCF
In a recent interview, Jeremy describes how we use computer simulations and neutron scattering experiments to understand domain motion in proteins.
Read the interview here: OLCF

Our mercury project has been covered in the latest episode of Horizons on BBC.
eremy gave the keynote speech at the Tschira Foundation 20th anniversary symposium in Heidelberg.
Watch here:
Keynote Speech

Derek Cashmnan, fomer postdoc in the Baudry group and now faculty at Tennessee Tech, has been awarded a TN-SCORE seed funding award together with Jerome Baudry to continue their work on protein:protein interactions in photosystems.

John Ossyra won the “Best Presentation” award for UTK LS507 for his research presentation on the configuration of Brownian Dynamics simulation models, which will be used to investigate unknown ribosome assembly reaction rates. Data from preliminary simulations were presented to demonstrate how structural models and parameter values are established for production quality runs. These reaction rates are essential for computational modeling of whole cells, and may help elucidate biologically relevant assembly pathways.
Congratulations John!

Jeremy has been interviewed by the Metro Pulse for a recent story on the UT-ORNL Governor’s Chair Program. In this article, some of the projects in our lab are introduced, too.
Read the article here: Metro Pulse
Our work on bioscavanger enzymes that can neutralize nerve agents, such as sarin, has been covered in several recent news articles.
Read here:
TIME
Tennessee Today
Health Medicine
Former graduate student Sally Ellingson from the Baudry Lab will continue her work on polypharmacology and supercomputer-based docking as a research faculty at the Markey Cancer Center and the Center for Computational Sciences of the University of Kentucky. She describes her work in a recent interview by NICS and a video for the ORNL.

Graduate students Xiaohu Hu and Jason Harris have been awarded with the 2014 Science Alliance graduate student award from UTK.
The Science Alliance has a mission to expand cooperative ventures in research with the Oak Ridge National Laboratory and recognizes achievements of UTK graduate students.
Congratulations!

In a recent press release, Loukas comments on his collaboration with researchers from the ORNL’s Environmental Sciences Division. In their study, they make use of supercomputer simulations and neutron reflectometry to analyze the structure of organic matter in soil systems.c
Amandeep’s research on lignin polymerization has been covered in a recent press release.In this study she uses density functional theory to show how specific monomers prevent the growth of lignin.
Read the research paper here: The Journal of Physical Chemistry B
In a recent interview, Loukas and Jeremy comment on their progress in the Biofuels project and the role supercomputing plays in that project.
Read the interview here: OLCF
A recent press release describes the work of Jerome Baudry in collaboration with the Zhulin lab at the UT/ORNL Joint Institute of Computational Sciences and with researchers from the University of Utah.
Their simulations and experiments on the signaling dimer of the E. coli Tsrchemoreceptor suggest a phenylalanine rotameric switch to play a key role in signal transduction.
Read on: OLCF
Read the research paper here: Nature Communications

Jerry Parks and Jeremy Smith of CMB are part of the Mercury SFA Team that won both the 2013 UT/Battelle Award for Scientific Achievement and the 2013 ORNL Director’s Award for Outstanding Team Achievement.
Congratulations!
Our work on understanding the pretreatment of lignocellulosic biomass by experimental techniques and supercomputing has been in the news. A recent article in Green Chemistry describing the underlying physical factors of this process features Loukas Petridis, Benjamin Lindner, and Jeremy Smith.
Read on: Oak Ridge Today
Read the research paper here: Green Chemistry

Graduate students Jing Zhou and Karan Kapoor have passed the GST Comprehensive exam (known as the “Prelim”) to become PhD candidates.
Congratulations to Jing and Karan
A new publication by Zheng Yi et al. appears on the cover of the next issue of The Journal of Chemical Physics. In this work, molecular dynamics simulations are analyzed with a Markov State Model.
Read on: The Journal of Chemical Physics
Computer chip maker Intel opens the Center for Parallel Computing at the UT/ORNL Joint Institute for Computational Sciences dedicated to the development of code for parallel calculations on supercomputers. Our project on optimizing the molecular dynamics software Gromacs is the first project to receive funding.
Read on: Oak Ridge Today
Sally Ellingson and Jeremy Smith comment on science funding for supercomputing on NPR.
Listen to the interview here: NPR
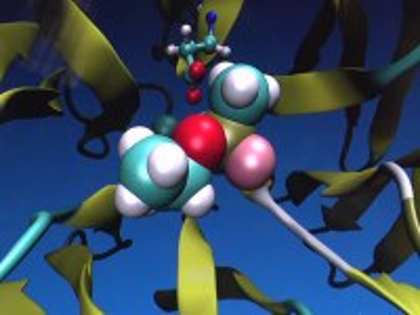
Our research project on the immunity to deadly chemical gases such as sarin has been covered in the ABC news. In collaboration with the Center for Structural and Molecular Biology, we work on engineering natural enzymes to convert dangerous nerve agents more efficiently. In order to provide protection from chemical weapons in a timely manner, more funding is needed.
A new publication by Yinglong Miao et al. appears on the cover of the next issue of The European Physical Journal E. In this work, the hydration dynamics of globular proteins were investigated with molecular dynamics simulations.
Read on: The European Journal of Physics E
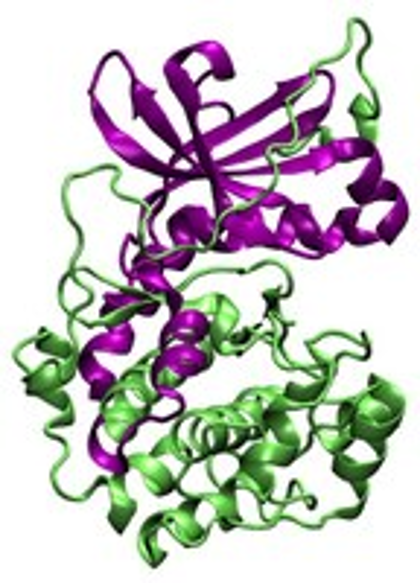
A proposal on simulating the structure and dynamics of protein kinase A by Loukas Petridis has received a 2013 ASCR Leadership Computing Challenge (ALCC) award from NERSC.
Congratulations Loukas!

Sally Ellingson has been awarded for her work in the Baudry lab with the American Chemical Society CCG award by the Computers in Chemistry division of the ACS and with the 2013 Science Alliance graduate student award from UTK.
The CCG award is given to no more than 10 graduate students every year to recognize the quality and significance of their research in the general field of computational (bio) chemistry.
The Science Alliance has a mission to expand cooperative ventures in research with the Oak Ridge National Laboratory and recognizes achievements of UTK graduate students.
Congratulations Sally!
A recent report in the Knoxville News Sentinelis about Jerry’s discovery on the transformation of mercury into its toxic organic form methylmercury. Together with Jeremy and other researchers from the University of Missouri, the University of Tennessee, and the Oak Ridge National Laboratory, he published the findings in a recent Science article.
Read the research paper here: Science
Liang’s work on protein flexibility is highlighted on the webpage of the American Physical Society. His latest interdisciplinary research is presented in which Liang worked with other researchers in order to distinguish the different types of protein motion.
Read on: APS
The cooperation of the Smith and Baudry labs with the research group of Nitin Jain from the Department of Biochemistry and Cellular and Molceular Biology has been mentioned in the news.
The three research groups made use of neutron scattering and NMR experiments along with computer simulations in order to understand the dynamics of Cytochrome P450 enzymes.
Read on: NICS

In an article for the ORNL Review,Jeremy describes the problems to overcome for the realization of biofuels and how our lab makes use of the world’s fastest supercomputers in combination with neutron-scattering experiments in order to tackle this task.

ORNL’s new supercomputer, Titan, has become available to users. With a peak performance of 20 petaflops, Jaguar’s successor is more powerful than any currently existing supercomputer.
Click on the following icons in order to watch, listen, and read recent interviews with Jeremy Smith and Jerome Baudry on how our research benefits from simulations run on Titan.
Jerome Baudry has been honored with the Outstanding Scholarship Award for Junior Faculty by the Department of Biochemistry, Cellular and Molecular Biology (BCMB). Jerome has joined BCMB in 2009 and his research group has been praised on more than one occasion for its scientific work.
Congratulations!
In a recent interview, Jeremy describes the role of supercomputers in the field of drug discovery.
Read on: HPC Wire
Sally Ellingson has won a NCSA travel Award to attend & present her work at the third European-U.S. Summer School on HPC Challenges in Computational Sciences. She will represent CMB together with Jerome Baudry and Jeremy Smith at this international conference which will take place on June 24-28, 2012, in Dublin, Ireland.
Congratulations to Sally!
Chelsea Knotts, a BCMB major and long-term undergrad in the lab of Jerome Baudry, has earned the highest student honor conferred by the University of Tennessee: The Torchbearer Award.
Chelsea, a Haslam Scholar and Lady Vols student athlete, has been awarded the Torchbearer mainly for her outstanding social work in the Knoxville community. She led hundreds of UT students in an effort to end chronic homelessness, particularly in the Fort Sanders neighborhood. She began a running group for homeless men and women, organized a 5K benefit run, and regularly befriends people who are without shelter and stability.
According to the Chancellor’s Honors website, this prestigious prize “is the highest honor the university gives to its students. Recognition as a Torchbearer reminds all students that those who bear the Torch of Enlightenment shadow themselves to give light to others”.
Great work Chelsea and congratulations!
The University of Tennessee’s Office of Research has honored William Hembree as the Scholar of the Week for his outstanding research achievements as an undergraduate student.
A chemistry major, William worked in the lab of Jerome Baudry at CMB and he recently published a paper on his research in the Journal of Physical Chemistry – something which he has also been complimented on by the Department of Chemistry in a recent interview.
UT faculty and students with exceptional accomplishments are usually awarded the QUEST – Scholar of the Week honor.
Congratulations to William!

The JICS/GRS workshop will be held on April 25-27, 2012 in Oak Ridge, Tennessee.
Organized by the Joint Institute for Computational Sciences at the ORNL and the German Research School for Simulation Sciences at Forschnugszentrum Juelich, this conference tries to explore how universities and government laboratories can work together to train the next generation of computational scientists.
Jerome Baudry is co-chair of this event and Sally Ellingson, Roland Schulz and Jeremy Smith each give a research talk.
For more information and registration, please go to the JICS/GRS homepage.
CBSB12, the sixth event in this international workshop series, will take place on June 3-5, 2012, in Knoxville, Tennessee.
Jerry Parks, Loukas Petridis and Jeremy Smith are co-organizers of this conference which brings together researchers from physics, chemistry, biology, and computer science to acquaint each other with current trends from computational biophysics to systems biology.
The registration deadline is April 15.
Find more information on the CBSB12 homepage.

Stay updated on CMB’s latest news, engage in scientific discussions and socialize with other followers on our facebook page.

As part of our developing relationship with the Shanghai Jiao Tong University, Qi Chen and Peng Lian, two graduate students from Jiao Tong, are currently visiting researchers at our lab for over one year. Peng works on the catalytic mechanism of cellulases by using QM/MM methods in the Guo lab and Qi deals with the HIV Integrase in the Cheng Lab.
We are pleased to have them work with us.

A recent ORNL story tip features our simulations on the characterization of the temperature dependence of the structure and dynamics of lignin polymers. The detailed characterization obtained provides insight, at the atomic level, both into processes relevant to biomass pretreatment for cellulosic ethanol production and general polymer coil-globule transition phenomena.
A 5-year report on our lab has been published. This booklet is packed full of awesome action, gorgeous graphics, heart-warming heroics, petrifying press releases, regal revelations and luscious lists. We hope you enjoy reading it.
Download it here: CMB (2006-2011)
In a recent press release on our work on lignocellulosic biomass, Loukas describes the simulation and experimental research done in our lab in order to characterize the structure of the polymer lignin and its role in the recalcitrance of biomass.
Read on: OLCF

The collaborative research effort by several ORNL groups on the detoxification of mercury has been in the news. Jeremy explains how supercomputer simulations performed in our lab contribute to the understanding of natural mercury transformation.
Our work on the hydration of a Cytochrome P450 enzyme has been in the news.
In this research project, Yinglong Miao and Jerome Baudry studied the active site hydration and
Our work on the hydration of a Cytochrome P450 enzyme has been in the news.
In this research project, Yinglong Miao and Jerome Baudry studied the active site hydration and diffusion of water molecules in the P450 enzyme. The protein hydration dynamics is relevant for the effectiveness of this drug-processing protein model. Further simulations are combined with neutron scattering experiments to understand dynamics of the protein enzyme.
Read on: NICS
In a recent interview with Volunteer TV,Jeremy describes how the Jaguar supercomputer helps to make our high-performance drug research possible.
Our work on combining neutron scattering experiments with molecular simulations in order to understand protein dynamics has been in the news. In this collaboration, three distinct and general classes, into which protein motion can be categorized, have been identified.
Read on: Science Daily
Our manuscript on the role of the two magnesium ions in DNA recognition by EcoRV restriction endonuclease was selected as cover of the Sept. 2nd 2011 issue of FEBS Letters.
Sally Ellingson, in the Baudry lab, was selected by the National Science Foundation among over 1,100 applicants to receive a scholarship to attend the 2011 Grace Hopper Celebration of Women in Computing Conference. Sally was also selected to give a presentation of her work in parallelization and cloud approaches to virtual docking.
Congratulations Sally!

Chelsea Knotts, BCMB major, Haslam Scholar, Lady Vols student athlete and long-term undergrad researcher in the Baudry Lab is in the news for her outstanding social work in the Knoxville community.
Great work Chelsea !!!!!!

A team of researchers from the Department of Energy’s BioEnergy Science Center has pinpointed a single, key gene in a microbe that could help streamline the production of biofuels from non-food sources.

The 2011 Conference in Computational Physics will be organized in Gatlinburg, Tennessee, USA, during the period 30th October — 3rd November 2011.
A press statement on the work on neurodegenerative diseases has been released. Our computer simulations have revealed how GSS-mutant proteins can misfold and result in plaque formation.
This project has been a joint effort with collaborators from Italy’s Università di Roma and University of L’Aquila including Dr. Isabella Daidone, a former visiting researcher at CMB.
Read on: Tennessee Today

A first of its kind combination of experiment and simulation at the Department of Energy’s Oak Ridge National Laboratory is providing a close-up look at the molecule that complicates next-generation biofuels.
The work published in Physical Review E was featured in an ORNL press release and picked up by several other websites: and publications:
http://www.physorg.com/news/2011-06-ornl-neutrons-simulations-reveal-bioenergy.html
and the June 2011, Volume 11, Issue 12 of the Materials 360(r) magazine.

Jerome Baudry has received the UT/BCMB Award for Junior Faculty Outstanding Teaching. Jerome is an assistant professor in the Biochemistry & Cellular and Molecular Biology department, at the University of Tennessee, and he and his group conduct their research in the UT/ORNL Center for Molecular Biophysics.
Dr. Baudry’s classes range from freshman general biology to senior undergraduate and graduate level structural biology and biophysics.
Congratulations Jerome!
Congratulations to Jeremy Spiers, former undergraduate in the Baudry lab, for his co-authorship on a paper in Biochemistry on anion-pi interactions in proteins in collaboration with Prof. Howell and Prof. Hinde. Other CMB co-authors are Prof. Jerome Baudry and Jason Harris, then rotation student in Prof’ Howell’s lab and now graduate student in Jerome’s group.

The Baudry lab has been awarded 50,000 Anton-hours on the new DESRES Anton supercomputer by NRBSC/PSC, for their NIH-funded Characterization of Dynamics Control of Chemotaxis Initiation project in collaboration with the group of Igor Zhulin. Multi-microseconds simulations of the dynamics of chemotaxis receptor’s signaling domain are on their way!

Chelsea Knotts, undergraduate in the Baudry lab and Haslam Scholar, has been awarded a competitive research stipend by The UT Knoxville Chancellor’s Office and Office of Research to pursue her research on environmental pollutants this summer with Jerome.
Congrats Chelsea!

Prof. Baudry and Sally Ellingson, graduate student in the Baudry lab, have been awarded an AWS in Education research grant for CPU time on Amazon’s proprietary Cloud architecture. This will allow the development of virtual docking on the Cloud.
Congrats Sally!
Prof. Jerome Baudry and Dr. Derek Cashman were the State Judges for the “Protein Modeling” event of the Science Olympiads that took place at the University of Tennessee, Knoxville. This is the first year that Protein Modeling is part of the tournament, that saw 17 of the top Tennessee High Schools’ science students competing in structural biology.
Read more about Tennessee Science Olympiad

Postdoctoral researcher Barbara Collignon has been elected to the NERSC Users Group Executive Board (NUGEX). She represents the Biological and Environmental Research division of DOE’s Office of Science at NUGEX. The purpose of this body is to promote the effective use of the high performance computing facilities at NERSC by providing advice and feedback.
Congratulations!
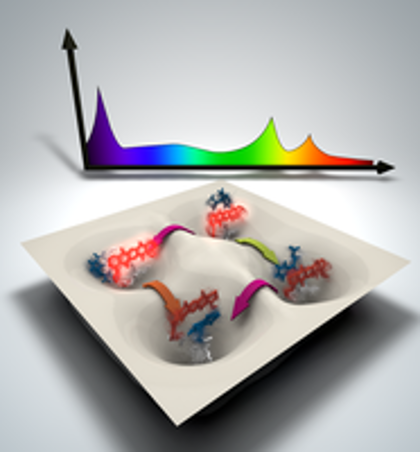
A theoretical technique developed at the Department of Energy’s Oak Ridge National Laboratory is bringing supercomputer simulations and experimental results closer together by identifying common “fingerprints.”
ORNL’s Jeremy Smith collaborated on devising a method — dynamical fingerprints –that reconciles the different signals between experiments and computer simulations to strengthen analyses of molecules in motion.
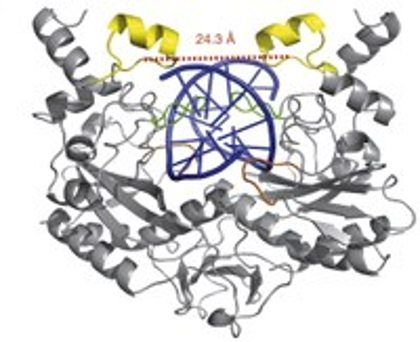
Our research is featured on the F100 Biologywebsite and the Naturally Selected blog of the The Scientist Magazine.
The mechanism of the progression in protein-DNA complex formation from separated molecules, to a nonspecific complex, to a specific complex has been viewed from protein-centric and more recently a DNA-centric perspective. The predominant protein-centric perspective in protein-DNA binding studies assumes that the protein drives the interaction. The mechanistic explanations for protein-DNA recognition usually arise from the analysis of the protein but this paper shows that the intrinsic flexibility of the DNA before the encounter is critical to specific complex formation.
EcoRV is a very well characterized endonuclease and the co-crystal structure reveals both DNA and protein conformational accommodations. In this case the DNA shows a 50 degree bend toward the major groove, which is possible due to a flexible TA in the center of the cognate sequence. The authors considered the consequences of having an AT instead. The analysis of the simulations suggested a three-step mechanism in the recognition of the cognate sequence and this was thwarted by the noncognate sequence. The process is found to be strongly influenced by the intrinsic bending propensities of the DNA free in solution before the protein approaches to form the nonspecific complex.
The Department of Energy has granted our research projects “Molecular Dynamics Simulations of Protein Dynamics” and “Virtual High-throughput Screening of Estrogenic Compounds” computation time at The National Energy Research Scientific Computing Center.

The University of Tennessee’s Office of Research has honored Jerome Baudry as Scholar of the Week for his group’s outstanding research work.
Congratulations!
Read more about QUEST – Scholar of the Week

The Department of Energy has awarded 30m processor hours on leadership computing facilities for the CMB’s research project “Cellulosic Ethanol: Simulation of Multicomponent Biomass System”.
Congratulations to Loukas Petridis, Xiaolin Cheng, and Jeremy Smith!
Read more about The INCITE Program
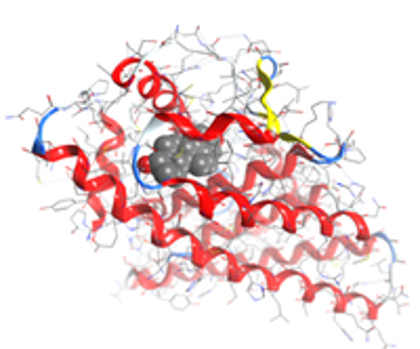
A quicker and cheaper technique to scan molecular databases developed at the Department of Energy’s Oak Ridge National Laboratory could put scientists on the fast track to developing new drug treatments.
A team led by Jerome Baudry of the University of Tennessee-ORNL Center for Molecular Biophysics adapted a widely used existing software to allow supercomputers such as ORNL’s Jaguar to sift through immense molecular databases and pinpoint chemical compounds as potential drug candidates.
Supercomputer simulations at the Department of Energy’s Oak Ridge National Laboratory are helping scientists unravel how nucleic acids could have contributed to the origins of life.
A research team led by Jeremy Smith, who directs ORNL’s Center for Molecular Biophysics and holds a Governor’s Chair at University of Tennessee, used molecular dynamics simulation to probe an organic chemical reaction that may have been important in the evolution of ribonucleic acids, or RNA, into early life forms. ….

Along with other UT scholars, Jeremy Smith presents the benefits the University of Tennessee provides to its students. This 2010 UT commercial will be aired during this season’s football and basketball games on TV.
Watch it on YouTube:
The University of Tennessee – Imagine a Better Future

Jeremy Smith has been invited to “Inside the Black Box – Science, Only Funnier”. In this interview on WREK Atlanta, 91.1 FM, he described the different aspects of biofuel research and how our group employs supercomputer simulations and neutron scattering experiments to analyze plant cell wall recalcitrance.

Two projects from the Center for Molecular Biophysics won 2010 DOE INCITE awards, giving us access to the world’s fastest computer. The two projects are:
1. “Cellulosic Ethanol: Physical Basis of Recalcitrance to Hydrolysis of Lignocellulosic
Biomass”; Principal-Investigator Jeremy C. Smith
2. “Interplay of AAA+ Molecular Machines, DNA Repair Enzymes, and Sliding Clamps at
the Replication Fork: A Multiscale Approach to Modeling Replisome Assembly and
Function”; Co-Investigator Xiaolin Cheng
About INCITE: The Innovative and Novel Computational Impact on Theory and Experiment (INCITE) program promotes cutting-edge research that can only be conducted with state-of-the-art supercomputers. The Leadership Computing Facilities (LCFs) at Argonne and Oak Ridge national laboratories, supported by the U.S. Department of Energy Office of Science, operate the program. The LCFs award sizeable allocations (typically tens of millions of processor hours per project) on powerful supercomputers to researchers from academia, government, and industry addressing grand challenges in science and engineering such as developing new energy solutions and gaining a better understanding of climate change resulting from energy use.
Mercury pollution is a persistent problem in the environment. Human activity has led to increasingly large accumulations of the toxic chemical, especially in waterways, where fish and shellfish tend to act as sponges for the heavy metal.
It’s that persistent and toxic nature that has flummoxed scientists for years in the quest to find ways to mitigate the dangers posed by the buildup of mercury in its most toxic form, methylmercury.
A new discovery by scientists at the University of Tennessee, Knoxville, and Oak Ridge National Laboratory, however, has shed new light on one of nature’s best mercury fighters: bacteria.
“Mercury pollution is a significant environmental problem,” said Jeremy Smith, a UT-ORNL Governor’s Chair and lead author of the new study. “That’s especially true for organisms at or near the top of the food chain, such as fish, shellfish, and ultimately, humans. But some bacteria seem to know how to break down the worst forms of it. Understanding how they do this is valuable information.” ……
Read on: Tennessee Today

Loukas Petridis and Benjamin Lindner present their work on atomistic modeling of lignocellulose at the Fifth Annual Georgia Glycoscience Symposium at the Complex Carbohydrate research Center in Athens, GA.

Tongye Shen from the Los Alamos National Lab is a new Assistant Professor at the BCMB Department of UT Knoxville and Group Leader at the Center for Molecular Biophysics. We are looking forward to working with him!

The ORNL Biophysics Summer School took place in Knoxville in the first week of August. During three days, scientists and students from around the country and from abroad presented their research and became acquainted with each other.
Xiaolin Cheng and Jerome Baudry from our group contributed to the organization of this conference. Jeremy Smith gave a talk on “Computer Simulation and Neutron Scattering in Biology”.
For viewing pictures of the event, please click here.

The push for alternative energy sources worldwide is leading to more advanced research in biofuels. Searching for new materials from which to produce such fuels is keeping researchers at Oak Ridge National Laboratory (ORNL) and the University of Tennessee (UT) busy.
A team led by Jeremy Smith, director of the ORNL Center for Molecular Biophysics and the UT-ORNL Governor’s Chair, will use the ORNL Jaguar and the UT Kraken supercomputers to run simulations that will help reveal the detailed workings of cellulose, a potential biofuel material.

Jeremy Smith and his fellow researchers have new fuel for understanding the barriers that keep biomass from becoming an economical source of ethanol.
Smith, director of the Center for Molecular Biophysics (CMB), an Oak Ridge National Laboratory-University of Tennessee (ORNL-UT) joint project, and his colleagues create computational models of lignocellulose. Those models could help us understand what makes this tough biomass component so difficult to break down into sugars for conversion to ethanol…. read on:
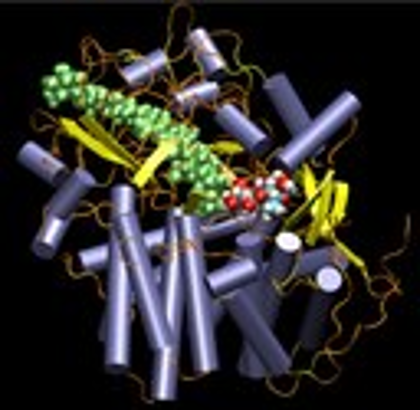
The anaerobic clostridium thermocellum produces an extracellular cellulase, Cellobiohydrolase CelS, which efficiently degrades cellulose by hydrolysing the glycosidic bonds. The catalytic domain of CelS in complex with substrate and product shows tunnel shaped substrate-binding region formed by the alpha helices in the protein. Enzymatic hydrolysis of the glycosidic bond takes place via general acid catalysis that requires two critical residues : a proton donor and a nucleophile/base. This process can happen with net retention or inversion of anomeric configuration. The structural analysis of CelS crystal indicates towards the inversion mechanism where the distance between the catalytic residues is significantly greater than that between the catalytic carboxylates of retaining enzymes. The hydrolysis in CelS possibly involves Glu87 as an acid to protonate the glycosidic oxygen atom and Tyr351 as a base to extract a proton from the nucleophilic water molecule that attacks the anomeric carbon atom.
Quantum mechanical/molecular mechanical (QM/MM) molecular dynamics and free energy simulations are performed on CelS and carbohydrate complex to understand the step by step mechanism for hydrolysis of glycosidic bond in cellulose and origin of product specificity.
by Jiancong Xu
The recalcitrance of lignocellulosic biomass to enzymatic hydrolysis is a bottleneck in cellulosic ethanol production. One promising avenue for overcoming biomass recalcitrance is to understand and modify the properties of bacterial cellulosomes.
Cellulosomes are large extracellular enzyme complexs that are produced by anaerobic bacteria and can efficiently break down plant cell wall polysaccharides, such as cellulose, hemicellulose and pectin into sugars. The cellulosome complex consists of various kinds of enzymes arranged around a scaffolding protein that does not exhibit catalytic activity but enables the complex to adhere to cellulose.
The organization of the cellulosome is mediated by high-affinity protein-protein interactions between Type-I cohesin domains within the scaffolding proteins and complementary Type-I dockerin domains carried by cellulosomal enzymes.
Computational modeling is essential to develop a full understanding of the assembly mechanism of the cellulosome and its ability in biomass digestion. Our initial goal is to explore the functional roles of specific domains and amino acid residues on the contact surface of the cohesin-dockerin complex by using molecular dynamics and free energy calculations.
ORNL has a long, distinguished history of catalysis research. With a diverse staff of world-class researchers and state-of-the-science facilities and instrumentation available on campus, ORNL addresses all aspects of catalysis research from very fundamental studies to specific applications. Our emphasis has been on heterogeneous catalysis and our researchers are also part of several national user centers at ORNL including the Center for Nanophase Materials Sciences, High Temperature Materials Laboratory, National Transportation Research Center, and Spallation Neutron Source. Thus, we host a large number of researchers from academia, industry, and other national laboratories who come to ORNL to solve their complex research problems.
Our focus has been on all aspects of heterogeneous catalysis including design and synthesis of novel catalysts, fundamental surface science, automotive emission catalysts, theoretical modeling, and characterization. These are exciting times for catalysis research when we have the ability to see nanocatalysts at the atomic level, and there are new techniques under development that will enable monitoring catalysts under operating conditions. We envision this catalyst center will continue to make broad contributions to fundamental advances in catalysis, as well as work with our industrial partners to facilitate introduction of new catalysts and technologies that meet the goal of efficient use of energy while reducing the impact on the environment.
-Michelle Buchanan, Associate Laboratory Director
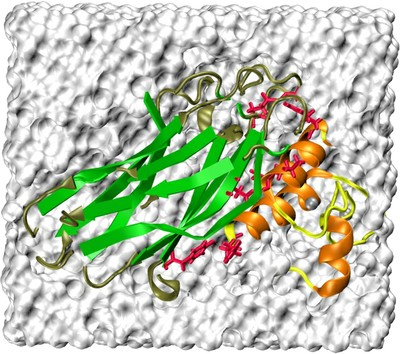
The molecular dynamics simulations, the product of an international collaboration led by ORNL researcher Jeremy Smith, are currently being pursued on the Cray XT4 Jaguar supercomputer. The simulations set the stage for neutron scattering experiments to test the theory by measuring protein motion.
Jeremy, who leads the ORNL Center for Molecular Biophysics and also holds a University of Tennessee-ORNL Governor’s Chair, collaborated with researchers from the University of Heidelberg in Germany. Their study has been accepted for publication in Physical Review Letters.
“The living cell is a network of proteins that talk to each other by interacting, sometimes transiently, sometimes for long periods of time. These interactions are important at every stage of cell function. Understanding the physical nature of these associations will help us to comprehend why they form and when,” Jeremy says.
The simulations performed by his team followed how a pair of interacting proteins move relative to each other as temperature increases.
Internal motions in proteins and many other materials undergo a rapid softening at a certain temperature, in a phenomenon called the “glass-to-liquid” or simply “glass” transition. The new simulations show that there is also a glass transition in the way the proteins in a pair, or ‘complex,’ move relative to each other.
At very low temperatures, around -200ºC, protein complexes are frozen stiff in a glassy state. But at around -40ºC, they suddenly free themselves up and behave like molecules in a liquid, diffusing randomly relative to each other, all while still remaining in touch, Jeremy says.
“The motion may in the future become measurable using specialized spectrometers at the Spallation Neutron Source, as neutron scattering has historically been a major technique for examining glass transition behavior,” Jeremy says. “Moreover, our team is wasting no time pushing forward with our calculations with a massive, 3.5-million-atom simulation of a thousand interacting proteins running on the ORNL Jaguar Cray XT4 supercomputer, even as the PRL article goes to press.”—B.C.
Article retrieved from ORNL Reporter

OAK RIDGE, Tenn., Sep. 27, 2007 — An international collaboration directed by an Oak Ridge National Laboratory researcher has performed the first-ever atomic-detail computer simulation of how proteins vibrate in a crystal.
Check out this link: https://phys.org/news/2007-09-interacting-protein-theory-awaits-neutron.html
September 17, 2007, KNOXVILLE — A team led by biophysicist Jeremy Smith of the University of Tennessee and Oak Ridge National Laboratory (ORNL) has taken a significant step toward unraveling the mystery of how proteins fold into unique, three-dimensional shapes.
A new culture at ORNL blends research with commercialization. There is no question. Science brought Jeremy Smith to Oak Ridge National Laboratory.
Jeremy’s Smith team uses supercomputer TITAN and the molecular dynamics coded GROMACS to run simulations explaining why lignin is such a problem for cost-effective ethanol. Along the way the refined a process for analyzing large-scale simulations using parallel analysis.
Click here for the full story.

Christy Franklin has been awarded the 2025 Biosciences Division Distinguished Achievement Award for Administrative Excellence! Christy has been our administrative

One of Jeremy’s ex graduate-students, Frauke Graeter, is taking up a Directorship at the Max Planck Institute for Polymer Research
We are proud to announce that Center for Molecular Biophysics, along with the Chemical Sciences Division, University of Tennessee, and

We are pleased to announce that former CMB Postdoc Vaidyanathan M.S. has accepted an offer from Oak Ridge National Laboratory

We are thrilled to announce that Madhulika Gupta, a former CMB Postdoc, has been nominated by the Anusandhan National Research

East Tennessee State University has recently granted Joshua Olaf Aggrey, a graduate student researcher at ETSU, the 2024 Appalachian Student

We are thrilled to have Mohan on our team at CMB, where he has already made significant contributions during his

Christy Franklin has been awarded the 2025 Biosciences Division Distinguished Achievement Award for Administrative Excellence! Christy has been our administrative

One of Jeremy’s ex graduate-students, Frauke Graeter, is taking up a Directorship at the Max Planck Institute for Polymer Research
We are proud to announce that Center for Molecular Biophysics, along with the Chemical Sciences Division, University of Tennessee, and

We are pleased to announce that former CMB Postdoc Vaidyanathan M.S. has accepted an offer from Oak Ridge National Laboratory

We are thrilled to announce that Madhulika Gupta, a former CMB Postdoc, has been nominated by the Anusandhan National Research

East Tennessee State University has recently granted Joshua Olaf Aggrey, a graduate student researcher at ETSU, the 2024 Appalachian Student

We are thrilled to have Mohan on our team at CMB, where he has already made significant contributions during his
Oak Ridge National Laboratory is managed by UT-Battelle LLC for the US Department of Energy